Next Generation Sequencing (NGS) has a crucial and pivotal role in medicine and drug development due to its ability to personalize treatment, enhance diagnostics, understand diseases at a genetic level, expedite drug discovery, and foster research advancements. This technology enables tailored treatments based on individual genetic profiles, accelerates disease diagnosis, uncovers disease mechanisms, identifies drug targets, and generates vast genetic data for comprehensive research, ultimately improving patient care and advancing drug development.
In 2015, cancer deaths accounted for 8.8 million of the total world death and the WHO also predicted an increase to 12 million deaths by 2030.(Fitzmaurice et al., 2017). However, the treatment of the various cancer types has been challenged with a lot of hurdles ranging from the insufficient identification of targets with high precision, the complexity of the anti-cancer drug discovery process, the tremendous cost incurred in the synthesis of drug, insufficient knowledge of the underlying molecular mechanisms and the absence of biomarkers that have been validated for the different tumour types(Yadav, et al 2014).
Next Generation Sequencing (NGS) has the ability however to eliminate these challenges and provide a better ground for early diagnosis as well as treatment of various cancer types. The role of NGS includes:
1. Precision Medicine:
Next Generation Sequencing enables and enhances the comprehensive research analysis of an individual’s genetic information, guiding tailored treatment plans based on the patient’s unique genetic makeup. This personalized approach enhances treatment efficacy and minimizes adverse effects. This in-depth understanding of a person’s genetic profile enables healthcare professionals to create personalized treatment plans specific to that individual’s genetic predispositions, providing more effective, targeted, and often safer treatments. It helps identify disease risk, choose the most suitable therapies, and predict responses to medications, thereby advancing the field of precision medicine by focusing on the unique genetic aspects of each patient for optimized healthcare outcomes (Strianese et al., 2020).
2.Diagnostic Improvements:
NGS revolutionizes disease diagnosis by offering accurate and rapid genetic profiling, aiding in early detection and treatment planning for various genetic disorders and cancers. These improvements also brought by NGS significantly enhance the accuracy, speed, and scope of disease identification, contributing to more effective and timely healthcare interventions.
3. Understanding Diseases:
NGS provides knowledge on the genetic basis of several diseases. This in turn, facilitates the identification of disease-causing genetic variations, improving the knowledge of the mechanisms of diseases and potential therapeutic targets.
4. Enabling Precision In Oncology:
Next-generation sequencing (NGS) provides a robust and efficient method for analyzing comprehensive genomic profiling. NGS-based technology facilitates the interrogation of multiple genes simultaneously, resulting in the rapid identification of targetable alterations. This is typically shown to be dependent on both tumour type and the stage of the disease (Clinical cancer genomic profiling. Nat Rev Genet. 2021). An example is the development of liquid biopsy which enables molecular analysis of circulating tumour DNA (Figure 1).
5. Drug Development:
The impact of NGS in medical and drug development is transformative, leading to more precise diagnostics, personalized treatments, targeted drug development, and a greater understanding of genetic aspects of diseases, ultimately improving patient outcomes and healthcare efficiency. It expedites drug discovery by identifying specific drug targets and allowing for the creation of more targeted therapies. NGS also assists in understanding drug responses, predicting adverse effects, and guiding the development of safer and more effective.
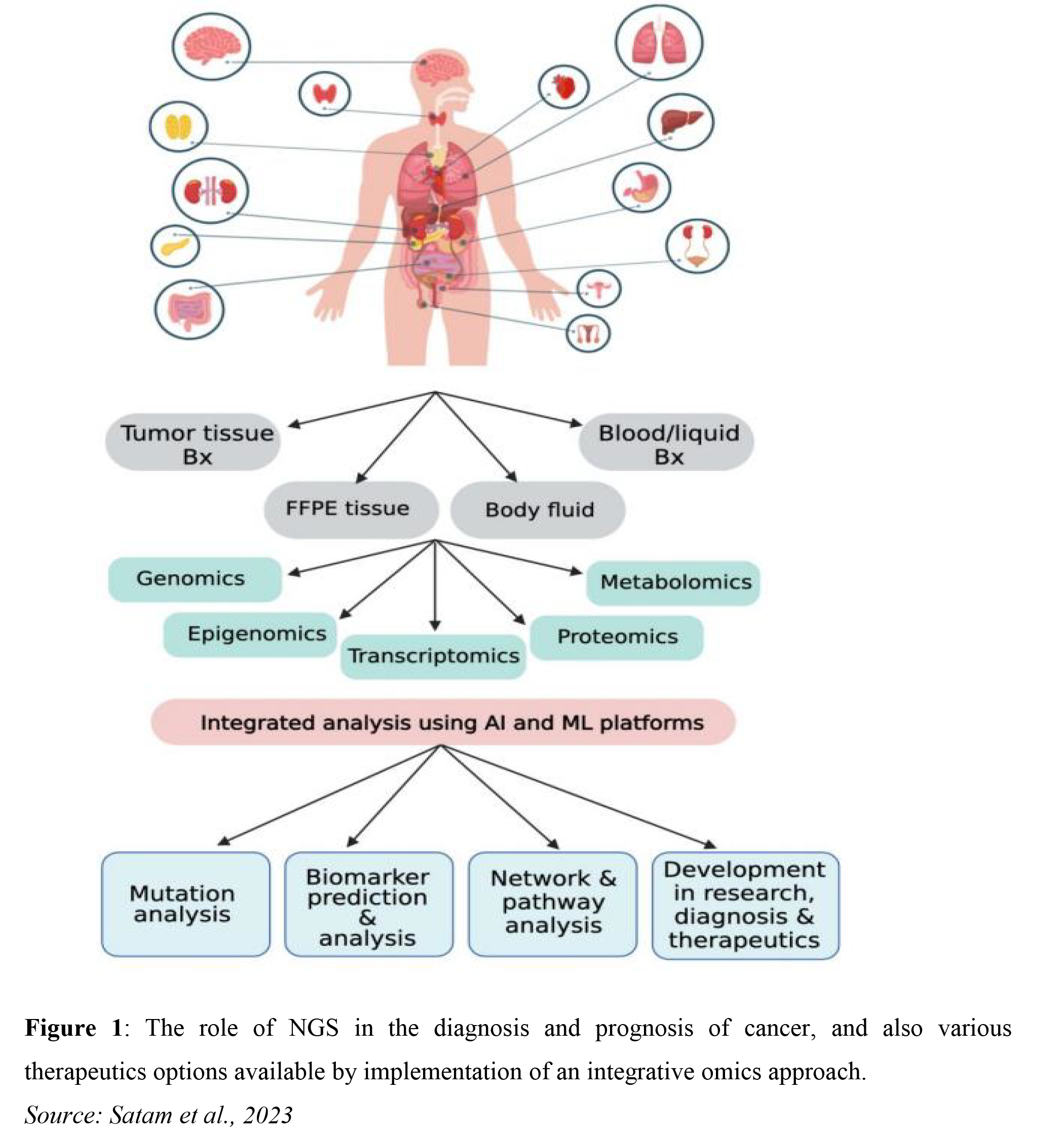
NGS accelerates research by generating vast amounts of genetic data, fostering a deeper understanding of genetic variations associated with diseases and drug responses. NGS has greatly impacted scientific research as a result of the high-throughput multiplexing. However, the strength of NGS in translational science is not solely based on the efficiency of the multiplexing but also on the various technologically advanced bioinformatic tools used for data curation and the various reference databases that help researchers, medical practitioners, and drug designers to understand the genetic basis of the disease (Satam, 2023).
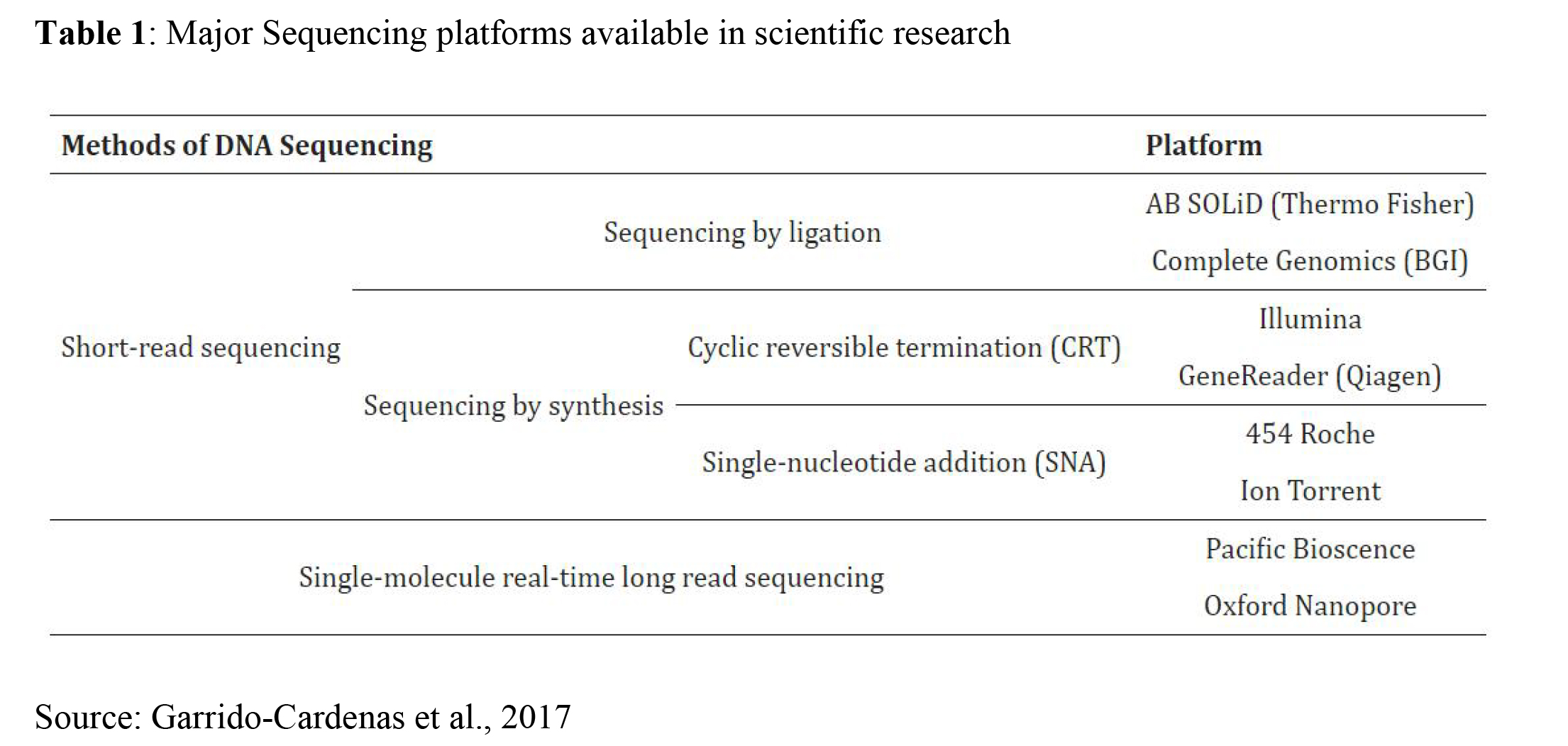
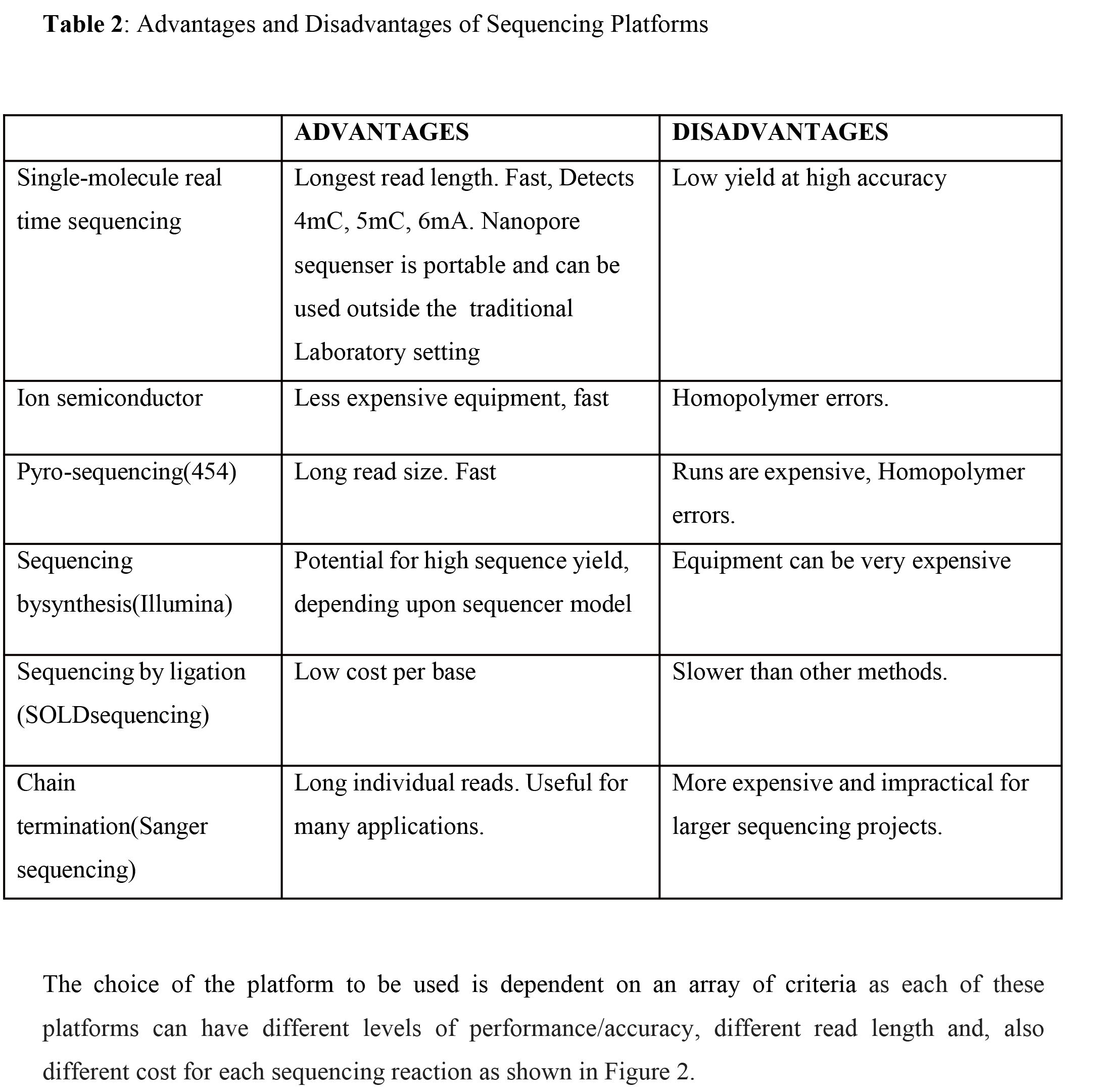
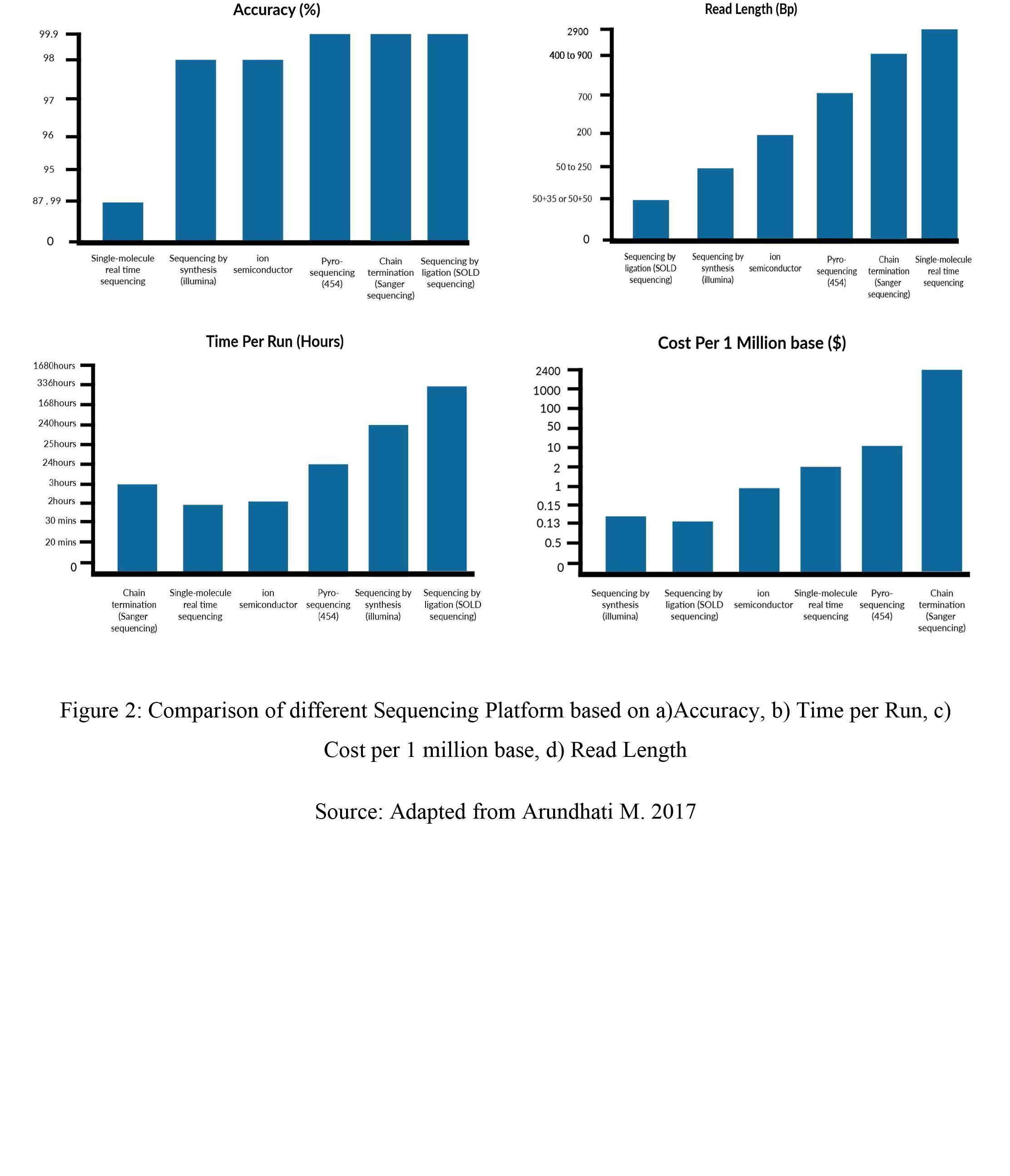
Several types/generations of NGS platforms exist that have helped in advancing scientific research as seen in Table 1. Table 2 also shows the advantages and disadvantages of each of these platforms.
NGS In Nigeria
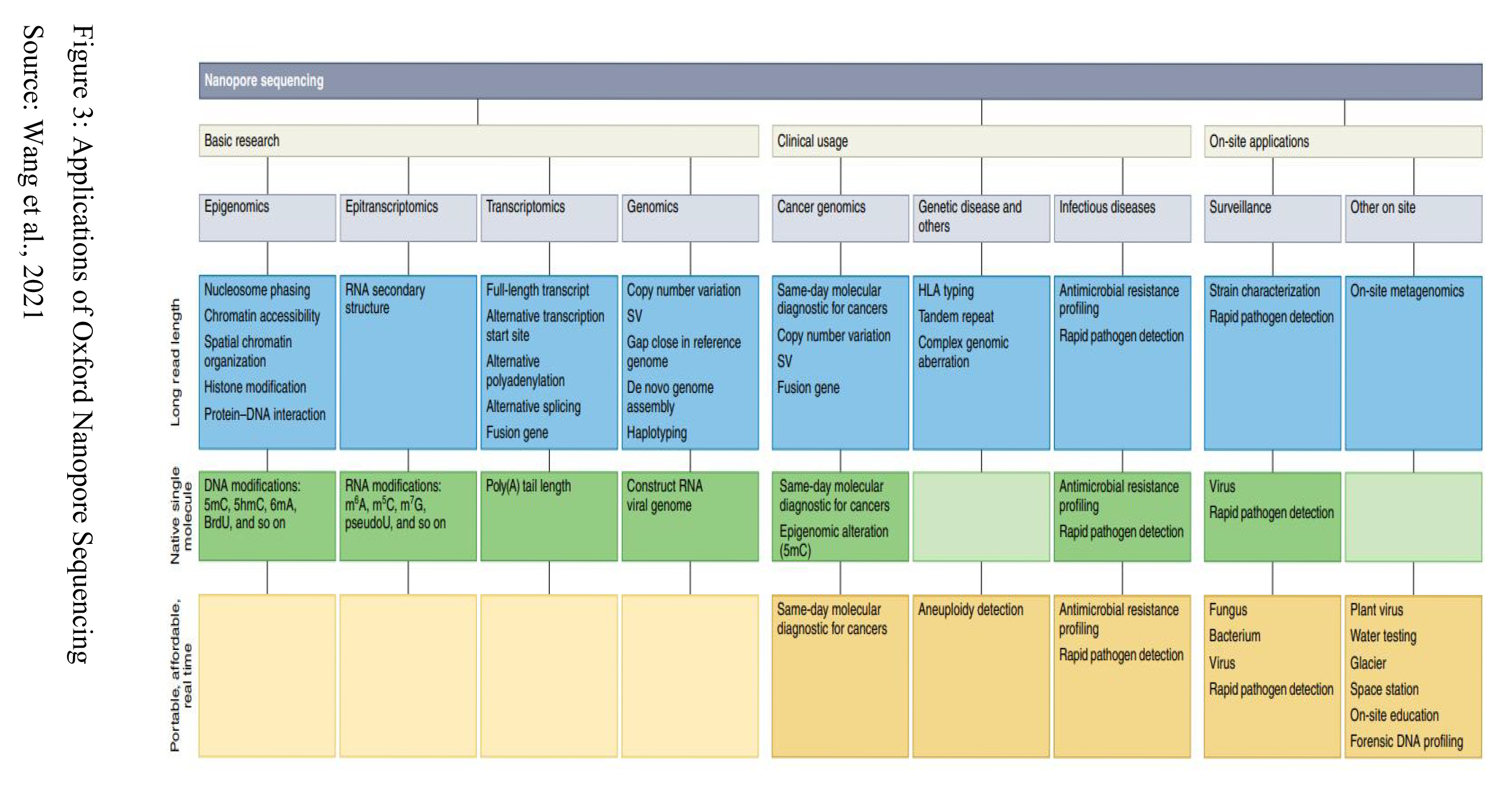
With regards to genetic service delivery in general, and NGS-based testing in particular, remains limited and extremely fragile in Nigeria and Africa at large. NGS-based diagnostics tests are not offered routinely to patients in Nigeria yet. The main barriers include limited existing infrastructure and processes, insufficient funding and lack of political support, and poorly structured health systems (Kamp et al., 2021). There is currently a misrepresentation of the African Genetic pool in the various reference databases in use. However, increasing diversity in databases with diverse African data will be a powerful tool to improve the diagnostic yield and the diagnostic accuracy for African and non- African undiagnosed diseases patients. Fortunately, the production of genomic data from African individuals has significantly increased over the last decade. This is because of the use of different sequencing platforms by few centers in Nigeria including the Nigerian Institute of Medical Research (DSBSEQ -D50), Inqaba Biotec (Sanger Sequencing using Applied Biosystems ABI 3500XL Genetic Analyser), NABDA, 54 gene (Illumina Novaseq 6000 and NextSeq 550Dx) and ACEGID (Illumina NovaSeq X Plus ). The Oxford Nanopore Technologies (ONT) has enabled many biomedical studies by providing ultralong reads from single DNA/RNA molecules in real time. Although the sequencing method is faced with limitations including relatively high error rates and the requirement for relatively high amounts of nucleic acid material, it is being employed in genome assembly, full-length transcript detection and base-pair modification detection and in more specialized areas, such as rapid clinical diagnoses and outbreak surveillance (Figure 3).The introduction of ONT into Nigeria clinical research will go a long way to solving various diseases faced by the Nigerian and African Population.
References
Fitzmaurice, C., Allen, C., Barber, R. M., et al; (2017). Global Burden of Disease Cancer Collaboration. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 3(4):524-548.
Yadav, M., Chatterji, S., Gupta, S.K., & Watal, G. (2014). Preliminary phytochemical screening of six medicinal plants used in traditional medicine. International Journal of Pharmacy and Pharmaceutical Sciences. 6. 539-542.
Strianese, O., Rizzo, F., Ciccarelli, M., Galasso, G., D’Agostino, Y., Salvati, A., Del Giudice, C., Tesorio, P., & Rusciano, M. R. (2020). Precision and Personalized Medicine: How Genomic Approach Improves the Management of Cardiovascular and Neurodegenerative Disease. Genes, 11(7), 747.
Satam, H., Joshi, K., Mangrolia, U., Waghoo, S., Zaidi, G., Rawool, S., Thakare, R. P., Banday, S., Mishra, A. K., Das, G., & Malonia, S. K. (2023). Next-Generation Sequencing Technology:Current Trends and Advancements. Biology, 12(7), 997.
Garrido-Cardenas, J. A., Garcia-Maroto, F., Alvarez-Bermejo, J. A., & Manzano-Agugliaro, F. (2017). DNA Sequencing Sensors: An Overview. Sensors (Basel, Switzerland), 17(3).
Arundhati M. (2017). Next Generation Sequencing. Availiable at : https://www.slideshare.net/arundhatimehta50/next-generation-sequencing-76687162. (Accessed 15/11/23).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y., & Au, K. F. (2021). Nanopore sequencing technology, bioinformatics and applications. Nature Biotechnology, 39(11), 1348. Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., & Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J Clin. 65(2):87-108.
Lee, H. &Tang, H. (2012). Next-generation sequencing technologies and fragment assembly algorithms. Methods Mol Biol. 855:155-74.
Levy, S. E., & Myers, R. M. (2016). Advancements in Next-Generation Sequencing. Annu Rev Genomics Hum Genet. 17:95-115.
Bao, S., Jiang, R., Kwan, W., Wang, B., Ma, X., & Song, Y.Q. (2011) Evaluation of next- generation sequencing software in mapping and assembly. J Hum Genet. 56(6):406-14.
Kamp, M., Krause, A., &Ramsay, M. (2021). Has translational genomics come of age in Africa? Hum Mol Genet. 30:R164–73
Sanmiguel P. (2011). Next-generation sequencing and potential applications in fungal genomics. Methods Mol Biol. 722:51-60